A first in prostate cancer research, scientists have applied a novel technology to unpack the genetic code of tumours.
A new avenue to differentiate between prostate cancers is being investigated by Vancouver Coastal Health Research Institute researchers Dr. Alexander Wyatt and Dr. Cameron Herberts. This research is one of the first to apply plasma cell-free chromatin immunoprecipitation sequencing (cfChIP-seq) to blood samples from advanced prostate cancer patients, offering new biological insights into why some prostate cancers are more aggressive than others.

“From a simple blood sample, we demonstrated that cfChIP-seq captures clear patient-to-patient variations in the epigenomes of their prostate cancers, partly explaining the divergent clinical characteristics of each patient’s disease,” states Wyatt.
“Understanding the biological determinants of different clinical patterns is a key step towards optimizing cancer care based on each patient’s individual needs.”
“We know that patients whose prostate cancer has spread to the liver can have different presentations and typically worse outcomes than individuals whose prostate cancer spreads only to their bones,” says Herberts. “These tumours can be very aggressive and difficult to treat.”
“Being able to anticipate the metastatic trajectory of a prostate cancer earlier on in the disease progression would lead to better-informed treatments tailored to a patient’s specific disease biology.”
An advantage of cfChIP-seq technology is that researchers can use it to identify new cancer features that could be targeted by precision medications. While the task of identifying prostate cancer subtypes was formerly akin to being shown millions of puzzle pieces and asked what the assembled work will look like, cfChIP-seq assembles more sections of the puzzle, offering additional clues to the final piece.
Unpacking the genetic code of prostate cancer cells
cfChIP-seq is a relatively new technique that enables study researchers to examine small fragments of tumour DNA, called circulating tumour DNA (ctDNA), in blood samples from patients with advanced prostate or bladder cancers from the B.C.-wide plasma cfDNA biobanking program.
When cancer cells die, they release their contents, including DNA, into the bloodstream. Researchers can use cfChIP-seq to measure specific molecular tags on nucleosome proteins — sections of DNA wrapped around histones — to learn which genes are turned on or off. Nucleosomes regulate how genetic information is packaged inside cancer cells and play a central part in the epigenome, which directs chemicals and proteins in the nucleus of the cell to attach to and mark genes to regulate which genes are active or inactive.
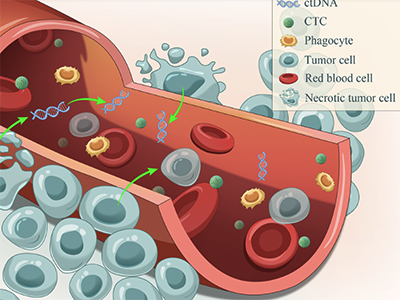
Using computational analyses of a tumour’s genetic code, scientists can retrace the development of cancer cells back to their beginnings, uncovering, for example, which genes the cancer cells were most dependent upon to form and spread.
“How genetic information is packaged inside cancer cells plays a key role in understanding its behaviour and clinical outcomes,” states Wyatt.
While historical chromatin immunoprecipitation sequencing (ChIP-seq) technology requires an invasive tissue or bone biopsy of a cancerous region to peer below the hood of nucleosomes, cfChIP-seq requires only a non-invasive blood sample to examine patients’ epigenomes via their ctDNA.

Researchers also found that some of the information from the cfChIP-seq profile of a patient with metastatic prostate cancer could be mapped to the other organs affected by their cancer, rather than originating from the tumour itself. “This suggests a potential role for cfChIP-seq in forecasting cancer-related organ damage before symptoms arise or monitoring recovery in real-time during treatment,” says Herberts. “While further research is needed to refine these applications, cfChIP-seq shows promise as a sensitive tool for tracking cancer’s impact on the body.”
“This research tool opens up a new dimension of biology for us to study,” Herberts adds. “Prior technology enabled us to know whether a gene had been mutated or not. However, the fact that cfChIP-seq can tell us whether a gene is turned on or off gives us greater granularity.”
“This new dimension of functional biology could help researchers understand how genes are working in relation to disease, potentially identifying new biomarkers of disease down the road.”
“Certain mutations mean that a patient could benefit from immunotherapy or some other form of targeted therapy,” Wyatt says. “Understanding not only how the genetic code of prostate cancer subtypes differs, but also how the genes are organized and packaged, is taking us to the next level of cell regulation research in prostate cancer.”
Along with applying cfChIP-seq to additional prostate cancers from a larger proportion of patients, Wyatt and Herberts are exploring additional clinical uses for the technology, including for the monitoring and treatment of other cancers.


